Information about Cloning Methods
Recombinational Approaches: att lambda phage approach (e.g. Invitrogen Gateway System)
 Description
Description
Invitrogen's Gateway (TM) system and other lambda phage (att) recombination site-containing plasmids can be used to add, replace or otherwise manipulate DNA using an enzyme that catalyzes site-specific recombination at att-type sites.
Learn More
To learn more about this method, please see the Invitrogen website www.invitrogen.com and appropriate references listed here:
- Festa F, Steel J, Bian X,and LaBaer, J. High-throughput cloning and expression library creation for functional proteomics. Proteomics (2013) 13:1381-99. PMID: 23457047
- Link AJ and LaBaer, J. High-Throughput Cloning of Open Reading Frames (ORFs): Assembling Large Sets of Expression Constructs. CSH Protocols (2008) Oct 1. PMID: 21356688
- Park J, and LaBaer J. Recombinational Cloning. Curent Protocols in Molecular Biology(2006) Chapter 3:Unit3.20. PMID: 18265386
- Landy A. Dynamic, structural and regulatory aspects of lambda site-specific recombination. Annual Review in Biochemistry (1989) 58:913-49. PMID: 2528323.
Advantages
Similar to the Cre/Lox approach, a significant advantage of the system is that an insert can be cloned into a single 'master' or 'entry' vector and readily sub-cloned into many different expression vectors without the need to digest with restriction enzymes, gel purify, ligate, etc (see Figure).
In the DNASU Plasmid Repository
Examples include the very many sequence-verified open reading frame (ORF) or cDNA clones in pDONR201 or pDONR221 shared with the repository by the the LaBaer Laboratory at Arizona State University's Biodesign Institute. Some of these clones are "closed" format such that the normal stop codon is present and others are "fusion" format in which the stop codon is replaced by a leu codon so that a C-terminal tag can be added.
Recombinational Approaches:Cre/Lox approach (e.g. Clontech Creator System)
Description
Clontech's Creator (TM) system and other LoxP site-containing plasmids can be used to add, replace or otherwise manipulate DNA using the Cre enzyme, which catalyzes site-specific recombination at LoxP sites. Please note that not every vector containing LoxP is appropriate for Cre/Lox-based cloning approaches. Sometimes the LoxP is present to make possible some experimental approach that is unrelated to cloning (for example, LoxP-containing constructs are sometime introduced into cells and then in vivo expression of Cre is used to bring about changes). In recent years, Clontech has discontinued its Creator products in lieu of new technologies, such as Flexi vector cloning, however you can still find the CRE recombinase from New England Biolabs.
Learn More
To learn more about this method, please see the Clontech website www.clontech.com and appropriate references, which include Hoess et al. (1982) P1 site-specific recombination: nucleotide sequence of the recombining sites PMID: 6954485.
Advantages
A significant advantage of the system is that an insert can be cloned into a single 'master' or 'entry' vector and readily sub-cloned into many different expression vectors without the need to digest with restriction enzymes, gel purify, ligate, etc.
In the DNASU Plasmid Repository
The many sequence-verified open reading frame (ORF) or cDNA clones in pDNR-Dual shared with the repository by the LaBaer Laboratory at Arizona State University's Biodesign Institute. Some of these clones are "closed" format such that the normal stop codon is present and others are "fusion" format in which the stop codon is replaced by a leu codon so that a C-terminal tag can be added. We have a number of plasmids and empty vectors available in DNASU that take advantage of Cre/Lox. You can find these using our plasmid feature search and selecting "LOXP" as the cloning system.
Restriction Digest Approaches: Standard RE Digests
Description
In standard restriction enzyme (RE) cloning, specific enzymes are used to cleave DNA at specific DNA sequences, and fragments with compatible ends (or blunt-ended fragments) are ligated to one another to form a circular plasmid, which can be transformed and propagated in bacteria. The method can be used to add, remove or otherwise manipulate DNA sequences.
Learn More
To learn more about this method, please consult a molecular biology textbook or protocol manual. New England Biolabs has additional tools and resources to use for restriction enzyme cloning.
Advantages
A significant advantage of RE cloning is that because it has been in use for many years, reagents and protocols are readily available to most researchers.
In the DNASU Plasmid Repository
Virtually any plasmid can be manipulated using restriction enzyme cloning methods. Particularly useful are those plasmids that contain a multiple cloning site (MCS), also called a polylinker. We have a number of plasmids and empty vectors available in DNASU that take advantage of restriction enzyme cloning. You can find these using our plasmid feature search and selecting "Multiple Cloning Site" as the cloning system.
Flexi cloning
Description
The Flexi® cloning system developed by Promega is a restriction enzyme-based system that uses rare restriction sites (SgfI and PmeI) to transfer a PCR product directionall into an expression vector. From this expression vector, the insert can be transferred into additional Flexi®-compatible expression vectors using restriction enzyme cloning
Learn More
Learn more about Flexi® Cloning from the Promega website and an article written by the developer of this system, Michael Slater. The Center for Eukaryotic Strucutral Genomics has also used the Flexi® Cloning system for high throughput cloning and protein production as part of the Protein Strucutre Initiatve and they have discussed their cloning pipeline on the SBKB Technology Portal and in the following publication:
- Blommel PG, Martin PA, Wrobel RL, Steffen E, Fox BG. High-efficiency single-step production of expression plasmids from cDNA clones using the Flexi Vector cloning system. Protein Expr. Purif. (2006) 47, 562-70. PMID: 16377204
Advantages.
The Flexi® cloning system is as efficient as Gateway cloning and has a lower occurance of mutations compared to Gateway cloning (Blommel et al.). This method is also faster, taking only 6-8 days to have sequence-verified Flexi® clones compared to 12 days in Gateway. That being said, the number of compatible Flexi® expression vectors is still fewer than compaitble Gateway expression vectors, which limits its overall flexibility.
In the DNASU Plasmid Repository
We have several Flexi compatible empty vectors available in DNASU. These include:
- pVP65K: A bacterial (E. coli) expression vector with an auto-cleaving N-terminal MBP tag that leaves an N-terminal 8xHis tagged protein
- pVP68K: A bacterial (E. coli) expression vector with N-terminal 8xHis and MBP tags.
- pVP33A: A bacterial (E. coli) expression vector with an N-terminal 8xHis and MBP tag cleavable by 3C or TEV protease
- pVP33K: A bacterial (E. coli) expression vector with an N-terminal 6xHis and MBP tag cleavable by 3C or TEV protease
- pVP56A: A bacterial (E. coli) expression vector with an N-terminal 8xHis and MBP tag cleavable by 3C or TEV protease
- pVP56K: A bacterial (E. coli) expression vector with an N-terminal 8xHis and MBP tag cleavable by 3C or TEV protease
- pEU-His-FV: A cell free (wheat germ) expression vector with an N-terminal 6xHis tag
- pEU-GST-FV: A cell free (wheat germ) expression vector with an N-terminal GST tag cleavable by 3C protease
PIPE cloning
Description
Polymerase Incomplete Primer Extension (PIPE) cloning was developed by the Joint Center for Structural Genomics (see publications below) as part of the Protein Structure Initiative. This method takes advantage of the fact that PCR reactions with incomplete primer extension result in single stranded DNA that can be annealed in a ligase-independent manner into complementary vectors (see Figure from Klock et al)
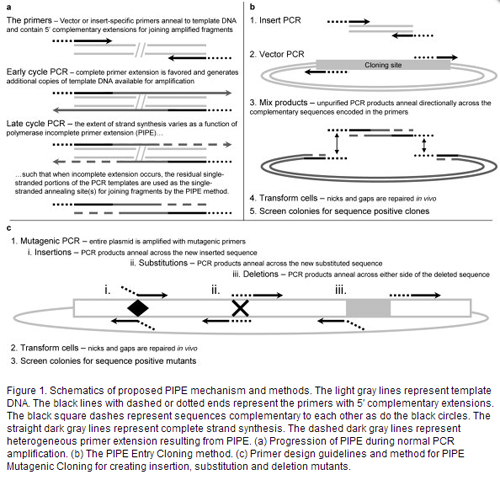
Learn More
Several PSI centers use PIPE Cloning for high throughput cloning and protein production as part of the Protein Strucutre Initiatve. Their cloning pipelines can be found by following the links to the SBKB Technology portal below and in the following publications:
- Joint Center for Structural Genomics
- Klock HE, Koesema EJ, Knuth MW, Lesley SA. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins. (2008) May 1;71(2):982-94. PMID: 18004753
- Klock HE, Lesley SA. The Polymerase Incomplete Primer Extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol Biol. (2009) 498:91-103. PMID: 18988020
- Mitochondrial Protein Partnership
- Partnership for Stem Cell Biology
- Kumar A, Möcklinghoff S, Yumoto F, Jaroszewski L, Farr CL, Grzechnik A, Nguyen P, Weichenberger CX, Chiu HJ, Klock HE, Elsliger MA, Deacon AM, Godzik A, Lesley SA, Conklin BR, Fletterick RJ, Wilson IA. Structure of a novel winged-helix like domain from human NFRKB protein. PLoS One (2012) 7:e43761. PMID: 22984442
- Transmembrane Protein Center
Advantages
The main advantage to PIPE cloning is that it provides a rapid method for creating large numbers of truncation mutations to screen for improved protein expression and crystallization. This method has both high cloning efficiencies and accelerated throughput without additional steps (such as a recombination recation) required for other methods (such as Ligation Independent Cloning or Gateway).
In the DNASU Plasmid Repository
We have a number of empty vectors create by the JCSG that are compatible with PIPE cloning. These can be found here. To search for all plasmids in DNASU that are in PIPE-compatible vectors, you can use our plasmid feature search and selecting "PIPE cloning" as the cloning system.
Ligation Independent Cloning
Description
Ligation Independent Cloning (LIC)is a fast and easy method for cloning that doesn't use restriction enzymes or DNA ligase. T4 polymerase is used to make specific 10-15 base overhangs on the target vector and the PCR product. The vector and insert are combined by mixing the two products together in the absence of ligase.
Learn More
Several PSI centers use LIC Cloning for high throughput cloning and protein production as part of the Protein Structure Initiative. Their cloning pipelines can be found by following the links to the SBKB Technology portal below and in the following publications:
- New York Consortium on Membrane Protein Structure
- A Ligation Independent Cloning protocol from NYCOMPS
- Midwest Center for Structural Genomics
- LIC Vectors
- Eschenfeldt WH, Lucy S, Millard CS, Joachimiak A, Mark ID. A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol. Biol. (2009) 498, 105-15. PMID: 18988021
- Nallamsetty S, Kapust RB, Tözsér J, Cherry S, Tropea JE, Copeland TD, Waugh DS. Efficient site-specific processing of fusion proteins by tobacco vein mottling virus protease in vivo and in vitro. Protein Expr Purif. (2004) 38, 108-15. PMID: 15477088
- Kim Y, Babnigg G, Jedrzejczak R, Eschenfeldt WH, Li H, Maltseva N, Hatzos-Skintges C, Gu M, Makowska-Grzyska M, Wu R, An H, Chhor G, Joachimiak A. High-throughput protein purification and quality assessment for crystallization. Methods. (2011) 55:12-28. PMID: 21907284
- Cleavable C-terminal His-tagged Vectors
- Eschenfeldt WH, Maltseva N, Stols L, Donnelly MI, Gu M, Nocek B, Tan K, Kim Y, Joachimiak A. Cleavable C-terminal His-tag vectors for structure determination. J. Struct. Funct. Genomics (2010) 11, 31-9. PMID: 20213425
- Kim Y, Babnigg G, Jedrzejczak R, Eschenfeldt WH, Li H, Maltseva N, Hatzos-Skintges C, Gu M, Makowska-Grzyska M, Wu R, An H, Chhor G, Joachimiak A. High-throughput protein purification and quality assessment for crystallization. Methods. (2011) 55:12-28. PMID:21907284
- Coexpression Vectors
- Stols L, Zhou M, Eschenfeldt WH, Millard CS, Abdullah J, Collart FR, Kim Y, Donnelly MI. New vectors for co-expression of proteins: Structure of Bacillus subtilis ScoAB obtained by high-throughput protocols. Protein Expr. Purif. (2007) 53, 396-403. PMID: 17363272
- Kim Y, Babnigg G, Jedrzejczak R, Eschenfeldt WH, Li H, Maltseva N, Hatzos-Skintges C, Gu M, Makowska-Grzyska M, Wu R, An H, Chhor G, Joachimiak A. High-throughput protein purification and quality assessment for crystallization. Methods. (2011) 55:12-28. PMID: 21907284
- LIC Vectors
- Center for High-Throughput Structural Biology
- Northeast Structural Genomics Consortium
Advantages
LIC is easy, fast and relatively inexpensive.
In the DNASU Plasmid Repository
Over 50 empty vectors in DNASU are compatible with Ligation Independent Cloning. A complete list can be found here. To search for all plasmids in DNASU that are in LIC-compatible vectors, you can use our plasmid feature search and selecting "Ligation Independent Cloning (LIC)" as the cloning system.

