Within the center, each step of our cloning, sequencing and distribution process is tracked by laboratory information management systems (LIMS) and facilitated by robots. Our processing pipeline is shown below.Roll over each image to see a zoomed-in view of the robot or process results.
Transformation and Colony Picking
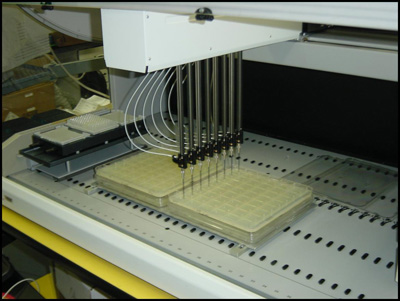
We receive plates of plasmids to be distributed through DNASU either as a 96-well plate of DNA or as glycerol stocks in phage resistant bacteria. When we receive DNA, we perform a transformation on the DNA in the entire 96-well plate and then plate the bacterial transformation on a patented 48-well agar plate using the FX liquid handling robot (see image). Individual colonies are picked into a deep well block using a K6 colony picker. Roll over the image to see the colonies growing on the 48-well plate after incubating overnight at 37 degrees Celsius.

Sanger Sequencing
After picking a colony and growing overnight in liquid culture, we copy the plate into a storage copy and a working copy. Using the FX liquid handling robot, we purify the DNA in high throughput for Sanger sequencing. Roll over the image to see a view of the chromatograph from the Sanger Sequencing machine.
Sequence Evaluation using ACE
Once sequenced, we use our Automated Clone Evaluation (ACE) software for sequence analysis. ACE uses artificial intelligence (AI) to analyze the raw sequence data by assembling the actual sequence of an insert into a contig and comparing it to its expected sequence. Each difference between actual and expected sequences is recorded as a discrepancy object including its position, bases, and sequence confidence. Acceptance and rejection are determined by counting the number of each discrepancy type and its confidence and comparing that against what is considered acceptable for a particular project. The discrepancies listed for a particular plasmid typically come from our analysis using ACE. Roll over the ACE screen shot for an example alignment.

Plasmid Storage in the Brooks BioStore Freezer
Once fully sequence verified, plasmid data is entered into DNASU and the samples are put into 2D barcoded tubes and entered into the Brooks BioStore Freezer Storage System. he freezer itself is 13,073 pounds fully loaded with samples. It is about 17.5 feet wide, 8.5 feet tall and 8 feet deep. It can store 855,000 tubes.The BioStore allows us to store glycerol stocks of our plasmid sample collection and easily retrieve and track these samples for distribution. Its primary benefit is its ability to “cherry pick” samples from within the collection. This saves time, safeguards our samples from cross-contamination and human error, and ensures that researchers will be able to receive materials from the repository more quickly. Roll over the freezer image for a close up of the picking robot.

